Data Inspector Module Setup
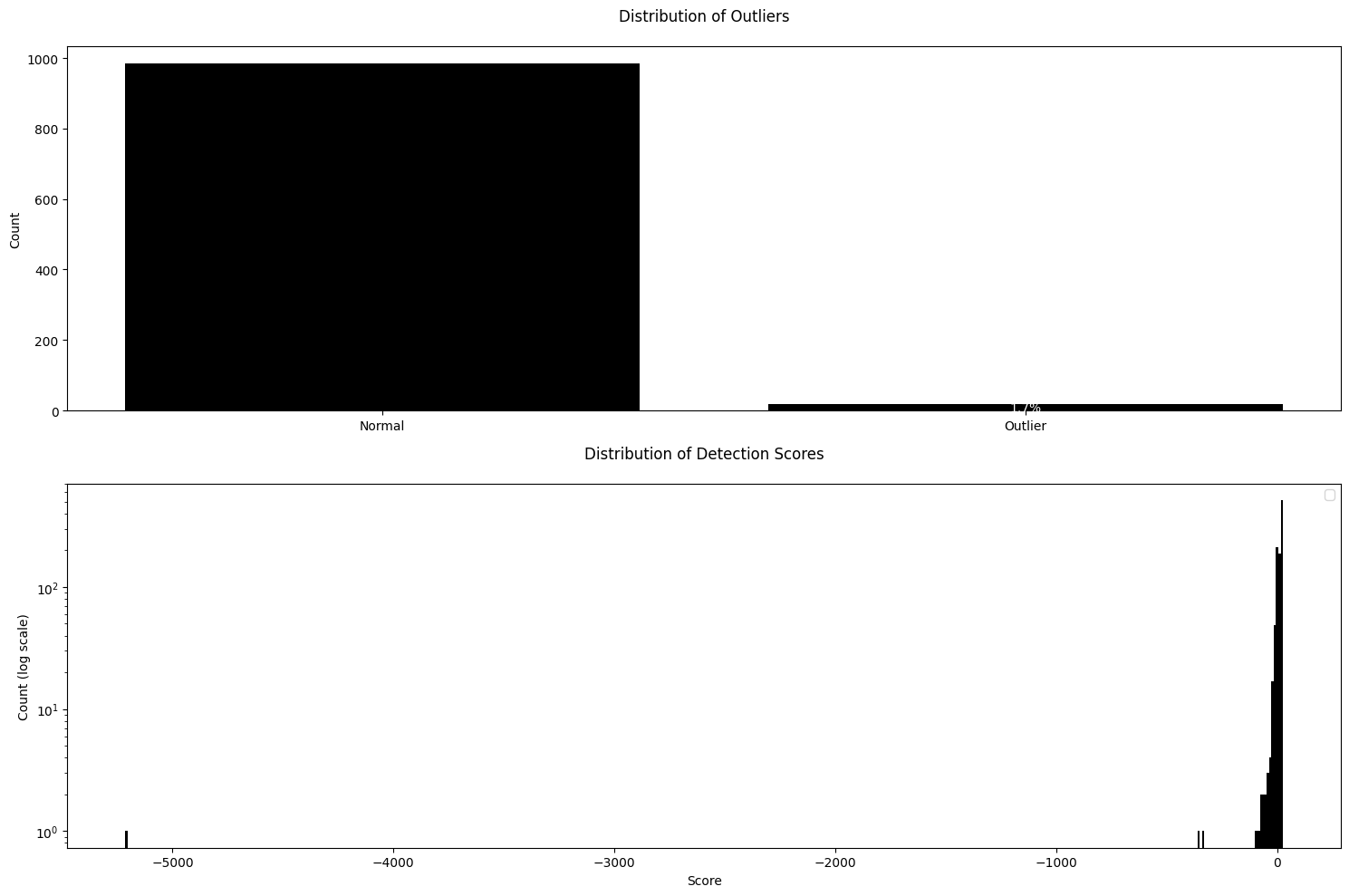
You are now ready to create an Outlier Detector — using the Reference dataset to obtain the typical distribution of visual features and applying that fitted model to the Inference dataset. The Obz AI package is designed to be highly modular and customizable. As a first step, we’ll import the specific feature extractors and the outlier detection algorithm you want to use.First Order Extractor and GMM Detector
FirstOrderExtractor is a straightforward and fast tool designed to extract first-order statistical features from images. These features summarize general properties of the pixel intensity values, such as mean, variance, skewness, etc. For example, they are useful for identifying images that are overly bright/dark or excessively variable in their intensities compared to the Reference dataset.
Note: First-order statistical features are invariant to the arrangement of pixels; in other words, they do not capture spatial relationships within the image.
At any point, you can view the list of features that FirstOrderExtractor computes by accessing its .feature_names attribute. This helps you to understand exactly which statistics are being extracted from your images.
GMMDetector is an outlier detection method that utilizes a Gaussian Mixture Model (GMM). To configure and use the GMMDetector, please follow the steps below:
extractors- Sequence of Extractor objects which process your data. Currently, only theFirstOrderExtractoris accepted.n_components- A number of Gaussian components for the mixture model. This controls the complexity of the model and how finely it can separate data clusters.outlier_quantile- Set the quantile threshold to determine what is considered an outlier. Data points falling below this quantile are classified as outliers.show_progress- If set toTrue, a progress bar will be displayed during feature extraction to visualize operation progress.
.fit method with a reference data. Ensure that the data you want to model comes in the form of a PyTorch DataLoader object.
.detect() method. This method returns a named tuple with:
img_features- extracted features for each image in the batch.outliers- boolean vector indicating if samples in the batch are outliers.

CLIP Extractor and PCA Reconstruction Loss Detector
GMMDetector applied on First Order Features provides a straightforward quantification that are invariant to the spatial arrangement of objects.
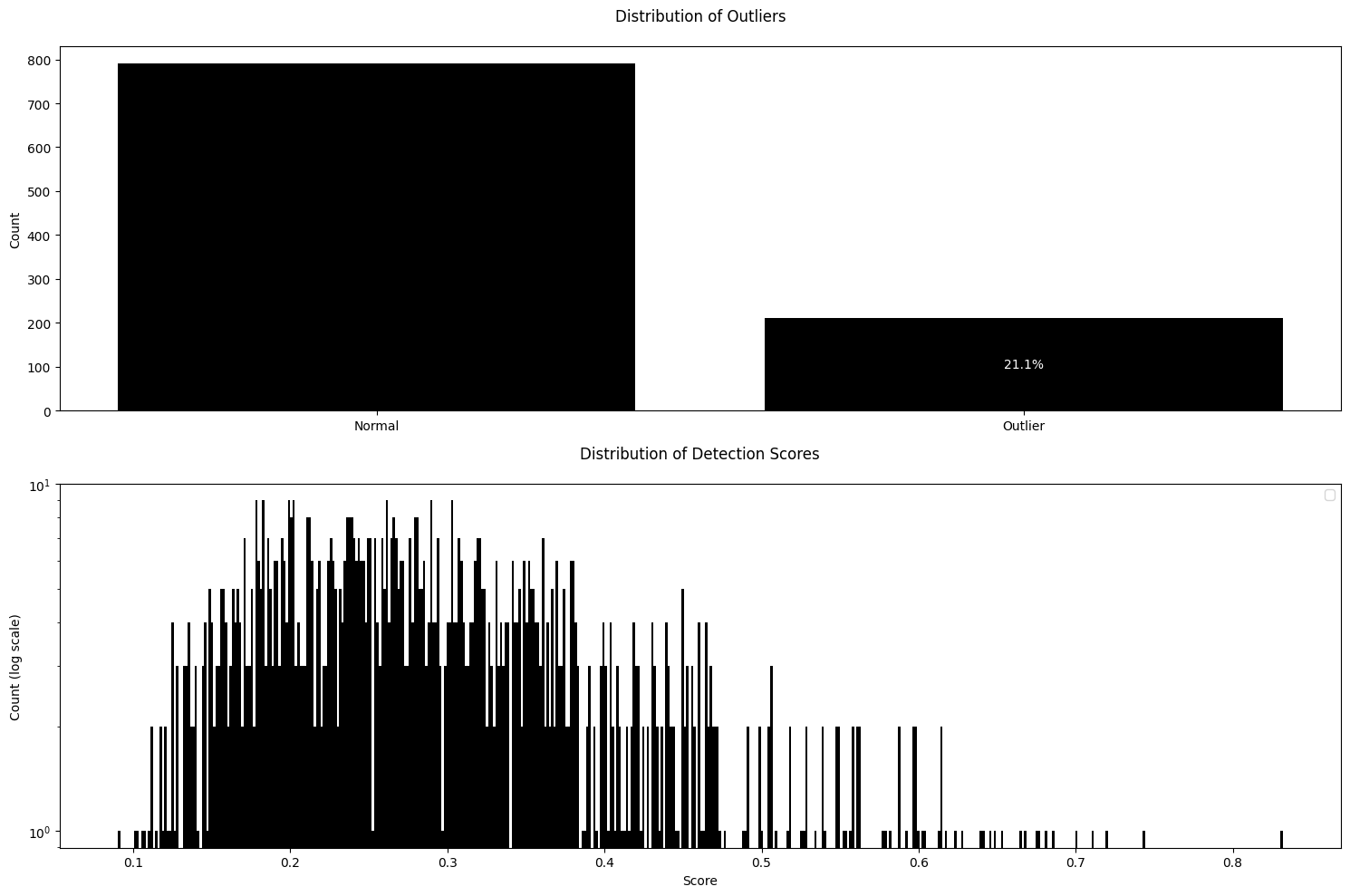
We provide more advanced outlier detection based on principal component analysis (PCA), which are suitable for high dimensional embedding features. Here, we look at how to use a deep learning model (CLIP; Contrastive Language-Image Pre-training) to extract embedding features that take account for spatial relationships. To handle this type of high dimensional features, we use PCAReconstructionLossDetector as an outlier detection algorithm.
There will an image schematic presentation of Reconstruction Loss idea.
To set up the PCAReconstructionLossDetector, you need to provide the following arguments:
- extractor: This is an already-instantiated extractor object that will be used to turn images into feature vectors. It is best to use an extractor that outputs features with high dimensions, such as
CLIPExtractor. - n_components: This is the number of principal components (PCs). In other words, it controls how many dimensions will be in the reduced latent space.
- outlier_quantile: This value is used to decide if an image should be considered an outlier. If the reconstruction loss (the difference between the original and reconstructed image in feature space) for an image is higher than this quantile, then the detector will classify that image as an outlier.
- show_progress: Set this to
Trueif you want to see a progress bar while features are being extracted. If you don’t want to see a progress bar, set it toFalse.
GMMDetector, PCAReconstructionLossDetector considers an unusually high value (e.g., reconstruction loss) to indicate evidence for an outlier.

Inspecting reference features
When you fit an Outlier Detector, it internally extracts and stores a set of reference features from your data. If you wish to access these reference features for inspection or further analysis, you can do so easily. Simply use the.return_reference_features() method on your detector instance. This method will return the reference features associated with the given detector.
Let’s see how to retrieve the reference features from a GMMDetector object:
.return_reference_2D_components()

