Setup
This tutorial introduces an overview of the Obz library which allows you to connect to the computer vision AI model. It provides a family of feature extraction and explanation methods. For the simplicity, we provide a pre-trained model and use an existing medical image dataset. We will go through the scenario step by step. Firstly, let’s start with doing some imports.Load a pre-trained model
Before you can monitor or visualize the workings of your model using our library, you first need to define the model you want to track. In this tutorial, we will use a Vision Transformer (ViT) model that has been previously trained and fine-tuned on lung nodules (LIDC) — meaning, the model has already learned to recognize patterns in medical images of lung nodules using a technique called self-supervised learning, specifically with the DINO backbone.Configure the ViT classifer based on DINO backbone
We are adding a binary classification head (see howtorch.nn.Linear) onto a DINO backbone.
Data Inspector
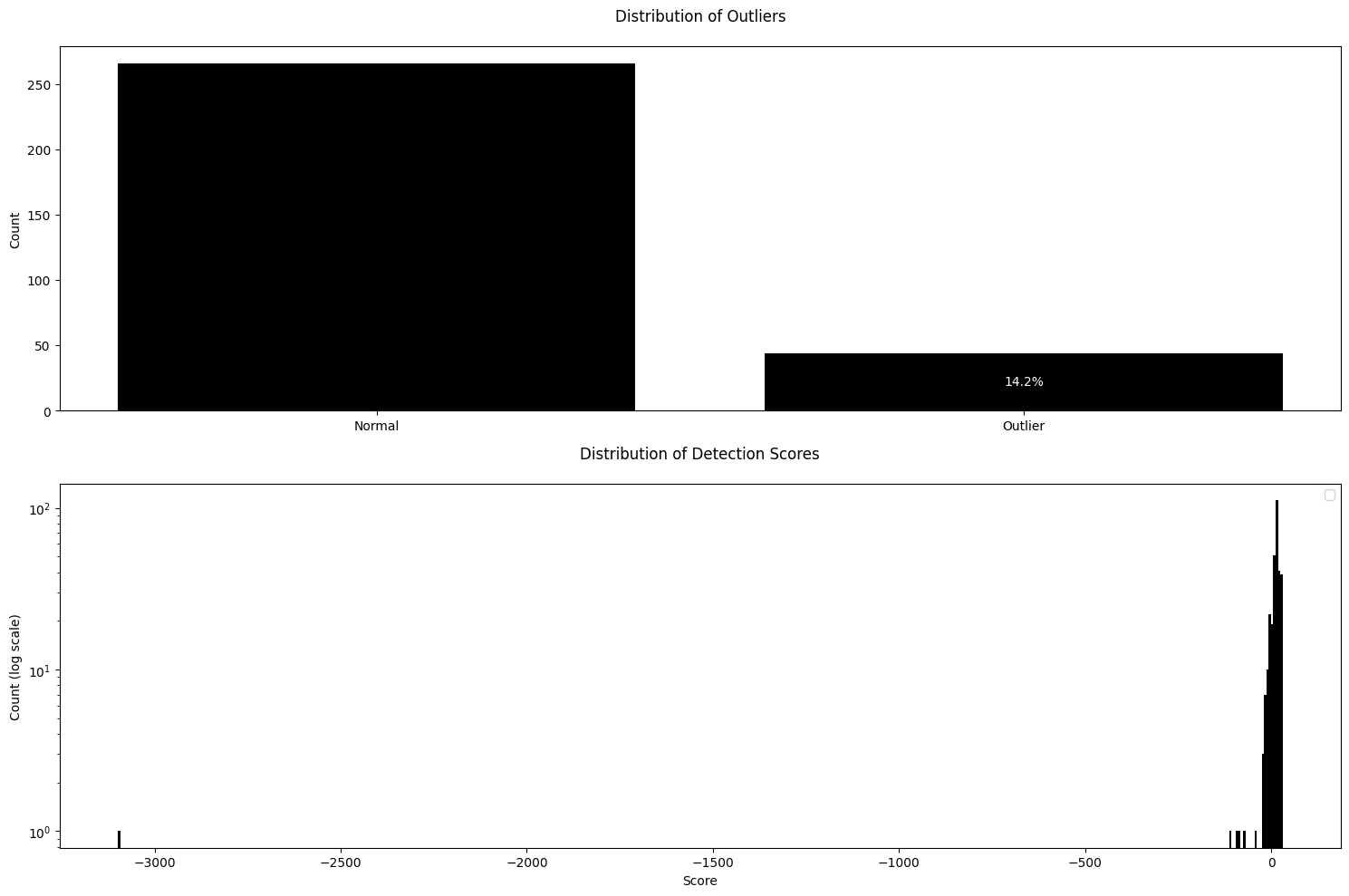
You are now ready to create an Outlier Detector — using the Reference dataset to obtain the typical distribution of visual features and applying that fitted model to the Inference dataset. The Obz AI package is designed to be highly modular and customizable. As a first step, we’ll import the specific feature extractors and the outlier detection algorithm you want to use.Extract First Order Features and build a GMM Detector
FirstOrderExtractor is a straightforward and fast tool designed to extract first-order statistical features from images. These features summarize general properties of the pixel intensity values, such as mean, variance, skewness, etc. For example, they are useful for identifying images that are overly bright/dark or excessively variable in their intensities compared to the Reference dataset.
Note: First-order statistical features are invariant to the arrangement of pixels; in other words, they do not capture spatial relationships within the image.
At any point, you can view the list of features that FirstOrderExtractor computes by accessing its .feature_names attribute. This helps you to understand exactly which statistics are being extracted from your images.
GMMDetector is an outlier detection method that utilizes a Gaussian Mixture Model (GMM). To configure and use the GMMDetector, please follow the steps below:
extractors- Sequence of Extractor objects which process your data. Currently, only theFirstOrderExtractoris accepted.n_components- A number of Gaussian components for the mixture model. This controls the complexity of the model and how finely it can separate data clusters.outlier_quantile- Set the quantile threshold to determine what is considered an outlier. Data points falling below this quantile are classified as outliers.show_progress- If set toTrue, a progress bar will be displayed during feature extraction to visualize operation progress.
.fit method with a reference data. Ensure that the data you want to model comes in the form of a PyTorch DataLoader object.
.detect() method. This method returns a named tuple with:
img_features- extracted features for each image in the batch.outliers- boolean vector indicating if samples in the batch are outliers.
NoduleMNIST.
XAI Module
XAI Module have components to both provide you with easy-to-use explainability tools and evaluation tools. Module consist of two major ingredients:- XAITool - Particular implementations of explainability methods.
- XAIEval - Evaluation methods for achieved explainability maps.
Predict the cancer status
Make the prediction for those 5 samples using the aforementioned MODELSetup XAITool and XAIEval
Computing importance scores with XAITool
cdam_tool- It is an excellent explainability method, highly discriminative with regards to the target class.smooth_grad_tool- Classical and simple XAI method.attention_tool- Classical way to inspect ViT like models.
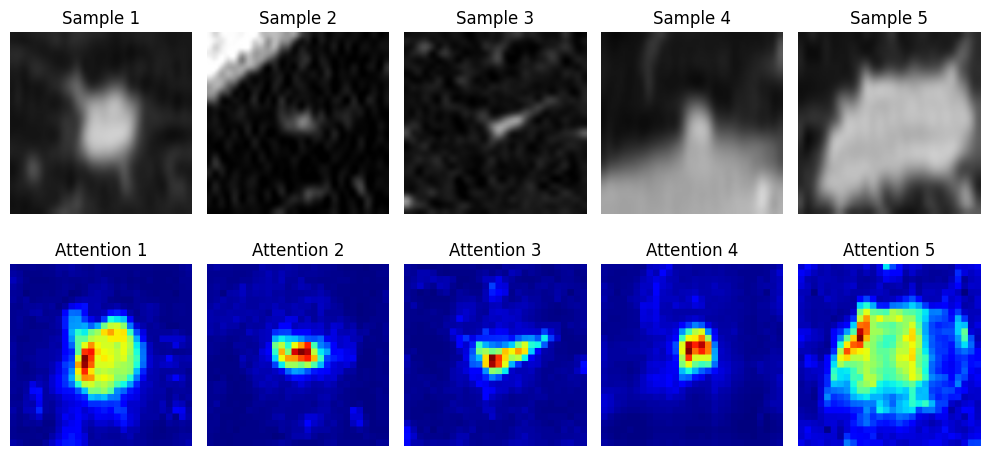
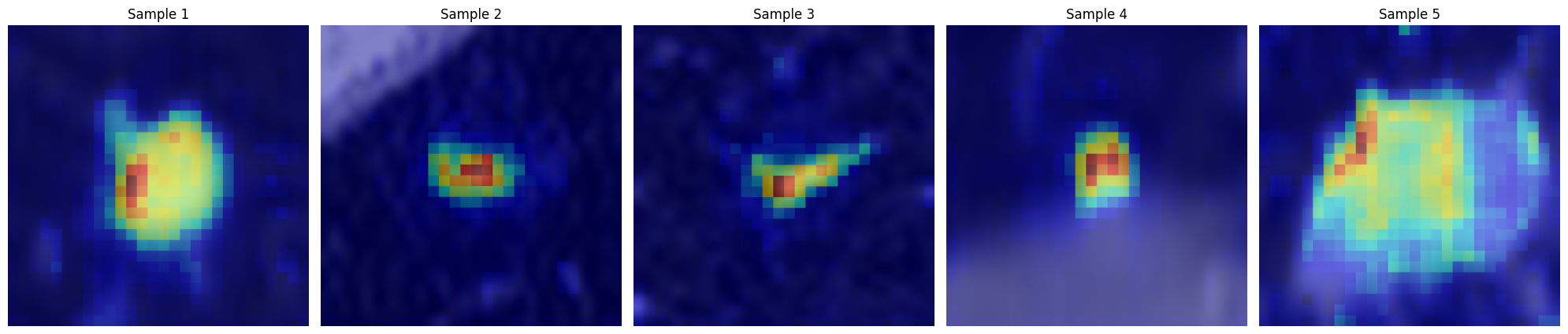
Generate and visualize CDAM maps
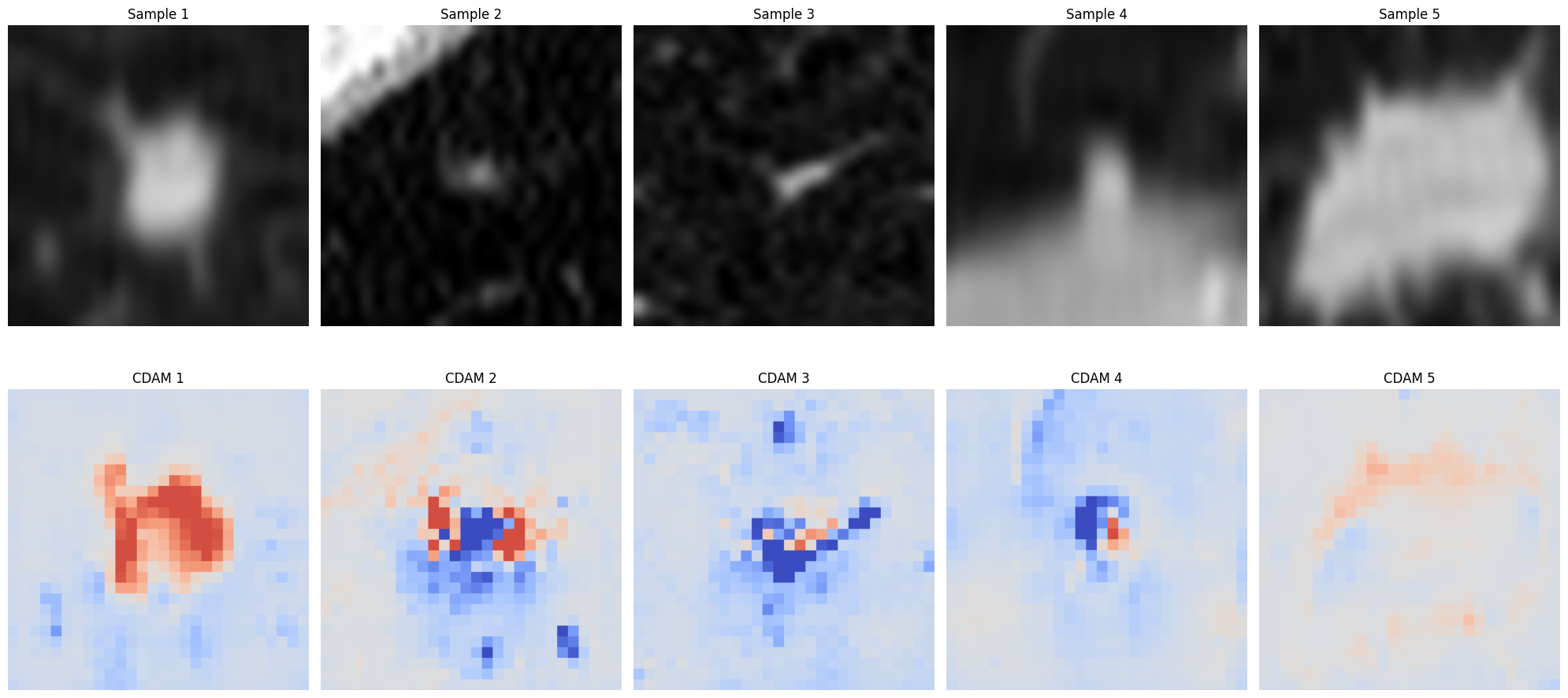
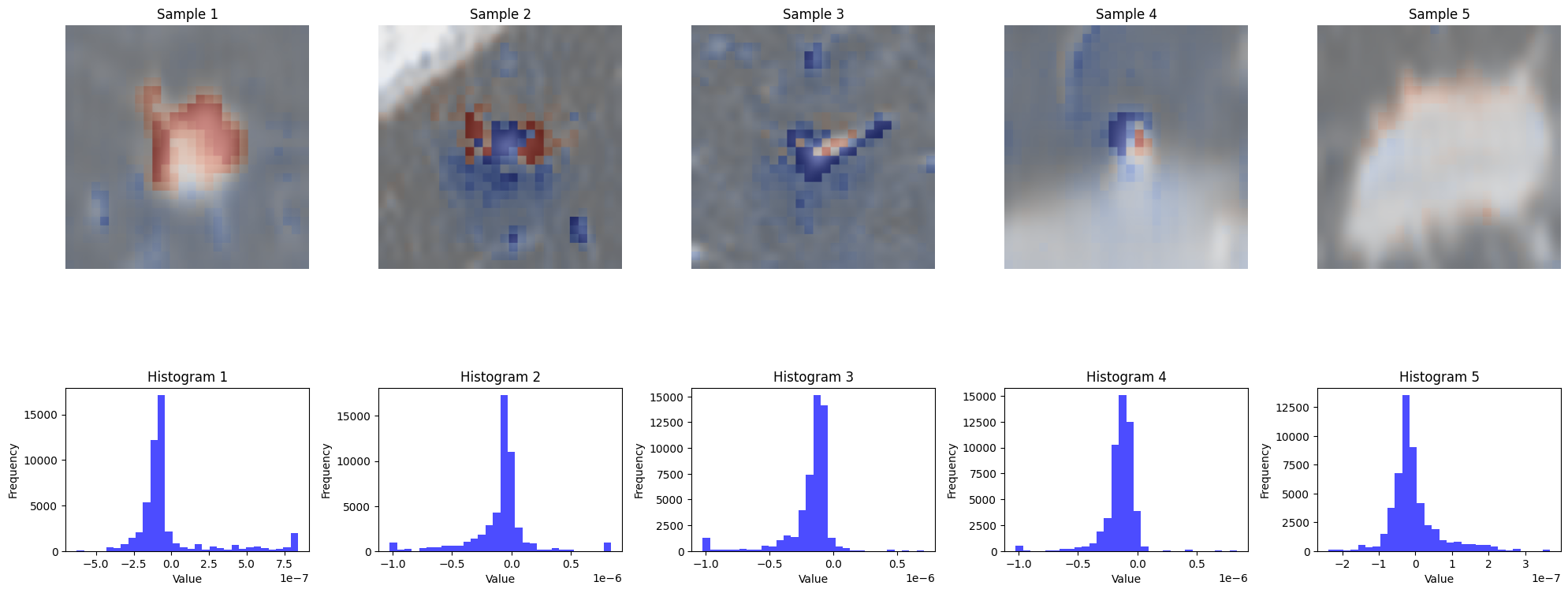
Evaluating XAI methods with XAIEval
There are several Explainable AI (XAI) methods available, each with their own advantages and limitations. Obz AI offers a set of evaluation tools to help assess the quality of XAI methods.
fidelity_tool measures how accurately a given XAI method reflects the model’s true decision process. It does this by systematically perturbing input features based on their importance scores and observing the resulting change in the model performance.
compactness_tool evaluates how sparse and concentrated the importance scores are. A more compact set of importance scores is often easier for humans to interpret, as it highlights the most relevant features in a concise manner.
By using these tools, you can better understand and compare the effectiveness and interpretability of different XAI approaches.
First, instantiate both evaluation methods:

